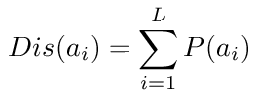
A great challenge in the proteomics and structural genomics era is to predict protein structure and function, including identification of those proteins that are partially or wholly unstructured. Disordered regions in proteins often contain short linear peptide motifs (e.g. SH3-ligands and targeting signals) that are important for protein function. Linear motifs are catalogued by ELM.
GlobPlot is a web service that allows the user to plot the tendency within the query protein for order/globularity and disorder. It successfully identifies inter-domain segments containing linear motifs, and also apparently ordered regions that do not contain any recognised domain. GlobPlot may be useful in domain hunting efforts. The plots indicate that instances of known domains may often contain additional N- or C- terminal segments that appear ordered. Thus GlobPlot may be of use in the design of constructs corresponding to globular proteins, as needed for many biochemical studies, particularly structural biology. GlobPlot has a pipeline interface - GlobPipe - for the advanced user to do whole proteome analysis.
GlobPlot has succesfully been used to:
- optimise constructs for expression, purification and crystallisation
- predict regions of enriched in linear motifs
- identify intrinsically disordered proteins (IDPs/IUPs)
The algorithm behind GlobPlot is shown below. We have changed it slightly from the originally published algorithm so it does not
include a logarithmic term anymore:
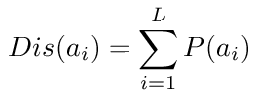
If you need further help please contact Rune Linding.